Since antibiotics and biocides have several target regions in bacteria, the most prevalent mechanisms for cross-resistance through non-specific activities such as efflux pumps and changes in cell wall characteristics (e.g., permeability reduction due to porin down regulation). [17] Based on that, in our study selected 11 bacterial isolates which shown higher antimicrobial resistance and evaluated through in vitro study against 12 various biocides on Muller hinton agar which includes disinfectants and antiseptics as shown in Table (2). Our results shown that the bacterial isolate revealed high resistance to Povidone-Iodine 10 % and climate disinfectant at percentage 81.82%, while Povidone-Scrub 7.5% and Spray disinfectant, at percent 90.91%. The best antiseptic used which shown highest effectiveness and none of the isolates (0%) shown resistance to it was Surgical soap, the rest of biocides revealed (100%) bacterial resistance as shown in Table (2). Surgical soap which contains Chlorhexidine gluconate 4%, Chlorhexidine digluconate (CHG), a cationic broad-spectrum antiseptic, disrupts bacterial membranes. As a result, the bacterial membrane has a negative charge that is stabilized by divalent cations (Mg2+ and Ca2+), which can be disrupted by chlorhexidine via (i) binding directly to the negative charge of the lipopolysaccharide (LPS) and (ii) displaced of divalent cations (LPS-stabilizing factor) which results in intracellular bacterial component precipitation. The usage of chlorhexidine corresponds with a reduced chlorhexidine susceptibility and a cross-resistance to antibiotics, particularly colistin, in P. aeruginosa. [18] Quaternry ammonium compounds (Qacs) are the main component of most biocides used so their resistance mechanisms are efflux pump as seen here. our bacterial isolates showed high resistance to biocide compared to other studies. This may be due to misuse or overuse of biocide and also the absence of execution of the instructions of the manufacturer, which results in subihibitory concentrations being applied, which results in bacteria becoming adapted to these biocides. Also, there are differences in geographical areas and bacterial strains, which have many resistance factors.
Our study revealed that out of Twenty isolates, 14 resistant isolates (70%) were EtBrCw-positive and 6 isolates (30%) were EtBrCw-intermediate as shown in Table (3) and Figure (1) which exhibited the method. This inducates that all isolates (100%) have efflux activity. This approach identifies overexpressed efflux systems that contribute to the MDR phenotype in bacteria. It uses EtBr as the pump substrate to verify the existence of an overexpressed efflux system in comparison to the intrinsic efflux activity of the identifying wild-type strain. [19] It is an easy, instrument-free approach that uses agar plates with increasing concentrations of ethidium bromide and is readily applicable to a standard clinical microbiology laboratory. [20] also the advantage of this method could be uses many isolates on one agar plates. Our results are higher than the study in Nigeria, were shown only (59%) of the isolates phenotypically revealed efflux pump activity. [21]
MexAB-OprM was the first RND pump observed to target several antibiotic classes and common wild-type bacteria, resulting in multidrug resistance. It is continuously expressed and enables the pumping of numerous antibiotics, dyes, biocides, and organic solvents out of the cell. Their antibiotics substrates include chloramphenicol, β-lactams, carbapenems, fluoroquinolones, tetracyclines, trimethoprim, and sulphonamides. [22, 23] The qac genes, particularly qacE and qacEΔ1, have been linked to resistance to biocides, particularly quaternary ammonium compounds. Biocides resistance has already been described in biocides and microbial resistance and is found in both clinical and environmental strains. [24]
Our results shown that 19 (95%) of isolates have mexB genes, while the qacE Δ1 gene was observed in all isolates (100%) and none of the isolates (0%) harboured the qacE gene as shown in Figures (2), (3), (4). Similar to study in Baghdad, Iraq was detected mexB efflux pumps gene in all isolates understudy, while study in Egypt was detected Mex B in 18 isolates (51.4%). [12, 25]
Our results shown the qacEΔ1 gene higher than the study in Baghdad, Iraq (80%) qacEΔ1 in P.aeruginosa, in Brazil the qacEΔ1 gene was detected 88% of the multiresistant isolates carried qacEΔ1 gene, in Turkey the qacE∆1 genes were determined in 50.7% (n=38), but the qacE gene in our result similar the same study in Baghdad, Iraq where, no any isolate carried qacE gene and disagreement with the same study in Turkey where, the qacE detected in 65.3% (n=49) and discovered the frequency of qacE and qacE∆1 genes in carbapenem resistant isolates was found to be significantly higher than in susceptible ones. [26, 27, 28] Also disagreement to study in Iran where Genomic detection of qacE and qacΔE1, showed that 111 (92.5%), and 21 (17.5%), out of 120 P. aeruginosa isolates harbor the qacE, qacΔE1. [4] These variation in the existence of the genes may be due to the differences in isolates of bacteria, types of hospitals, procedures of administration of antibiotics and application of biocides in the hospitals, and may be these genes are plasmid encoded.
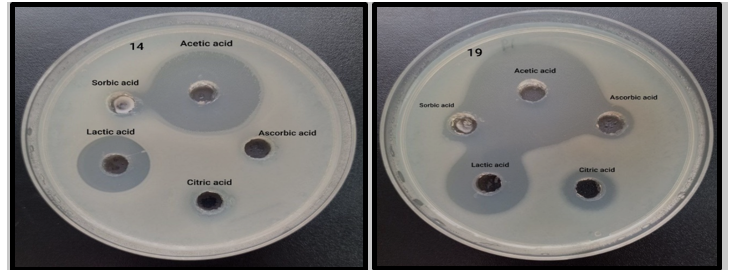
One of the worldwide health challenges is the rising prevalence of biocide resistance and the possibility of cross-resistance to certain antibiotics, which can lead to hospital-acquired infections and inefficient therapies. [29] the results of organic acids revealed that acetic acid and lactic acid have effective antimicrobial activity at a concentration 1.2% and 0.73% respectively, while citric acid, ascorbic acid and sorbic acid revealed antimicrobial activity against one isolate at concentration 0.24% as shown in Table (4) and Figure (5). So these concentration of acetic acid and lactic acid can be used as antiseptics and disinfectants and can be raised the concentration of others and testing antimicrobial activity. Energy competition, bacterial outer membrane permeabilization, increasing intracellular osmotic pressure, and suppression of biomolecule synthesis are some of the molecular mechanisms behind acid inhibition. Undissociated organic acids are lipid-soluble and can penetrate the cell through free diffusion. They dissociate to produce acid ions (ROO−) and protons (H+). The buildup of H+ in the cytosol contributes to cytoplasmic acidification, leading to rupture of cytoplasmic membrane, leakage of precursor and growth factor which result inhibiting metabolism and growth, acid ion block DNA synthesis, and denaturation of the important enzymes. [30]
Other study in India find out that 1% of acetic acid used as topical antiseptics on the burns to prevent and treat infections that have not responded to traditional therapy, including oral or injectable antibiotics and local wound care with hydrogen peroxide and betadine and the period of treatment required to eradicate MDR P. aeruginosa from the chronic burns was 4.5 days. [31] acetic acid at a concentration of 5% reveal an antibacterial and antifungal activity when tested on surfaces showed a complete reduction for P. aeruginosa, E. coli, S. aureus, E. hirae, A. brasiliensis and C. albicans. [32] Study found that acetic acid (vinegar) effectively kills M. tuberculosis after 30 min of exposure to a 6% acetic acid solution. [33] A high concentration of AA (>0.156% v/v) could be used to sterilize biofilm-prone surgical instruments, disinfect hospitals, and treat external wounds. A low concentration of AA (0.00975-0.039% v/v) can be used as an anti-virulence agent for adjuvant treatment of colistin-resistant P. aeruginosa, pH of AA below 4.76 was effective against P. aeruginosa biofilms. [34] These differences in concentration may be due to the nature of the cell wall characteristics of bacteria and the nature of surfaces dealing with it biotics or abiotics because environmental condition often effect on the effective concentration. lactic acid (LA) is a naturally occurring weak organic acid that is considered eco-friendly and recognized as safe. LA is used as a spray to decontaminate areas. [35]